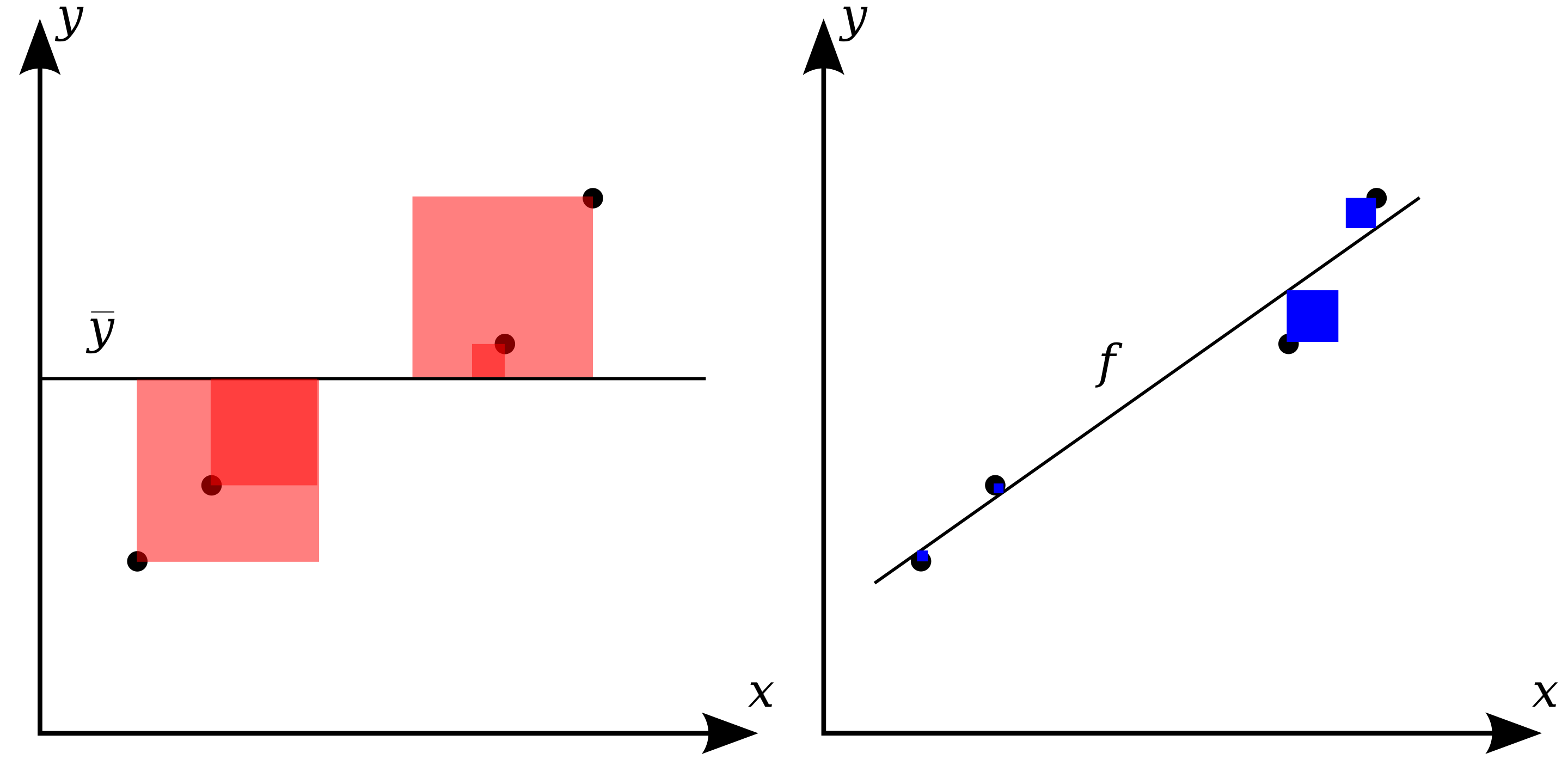
In statistics, the coefficient of determination, denoted \(R^2\) or \(r^2\) is the proportion of the variation in the observed variable (y) that is predictable from the explanatory variables (X). It is a performance metric for linear regression.
For given predictions \(\hat{y}_i\) and true labels \(y_i\), the \(R^2\) is:
\[R^2=1-\frac{SS_{Residual}}{SS_{Total}}=\frac{SS_{Explained}}{SS_{Total}}\]Where:
And law of total variance: \(SS_{Total}=SS_{Explained}+SS_{Residual}\).
The following graphics from Wikipedia shows a visual interpretation:

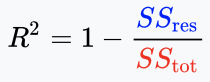
See:
For given predictions \(\hat{y}_i\) and true labels \(y_i\), the RMSE (or root mean square deviation - RMSD) loss is:
\[RMSE = \sqrt{\frac{\sum_{i=1}^n (y_i - \hat{y}_i)^2}{n}}\]See:
For given predictions \(\hat{y}_i\) and true labels \(y_i\), the MAE is:
\[MAE = \frac{\sum_{i=1}^n \vert y_i - \hat{y}_i \vert}{n}\]See:
Here is a representation of a confusion matrix:
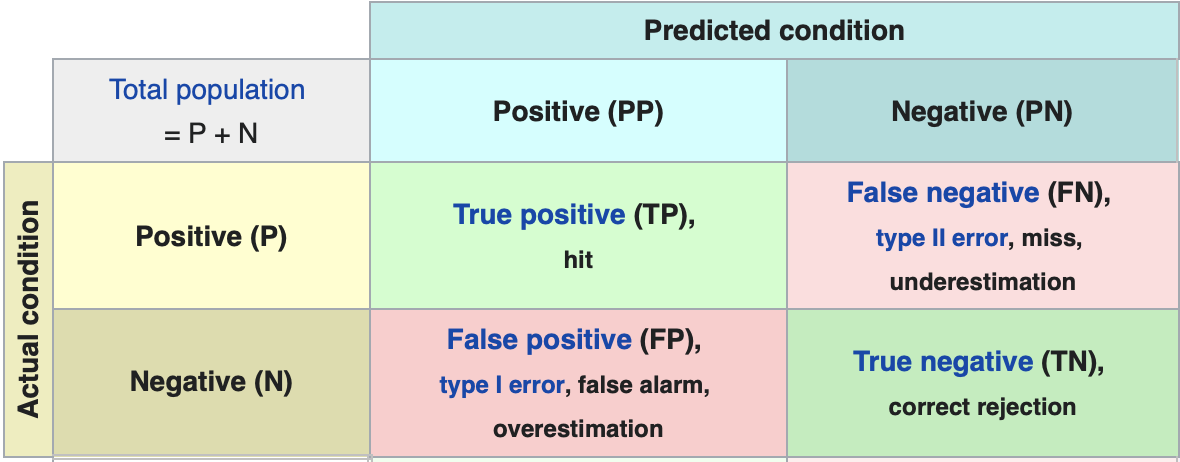
Where:
Precision measures how the model is accurate for the positive predictions:
\[Precision=\frac{TP}{TP+FP}\]Recall measures the percentage of the positive population that was detected positive:
\[Recall=\frac{TP}{TP+FN}=\frac{TP}{P}\]False Positive Rate measures the percentage of the negative population that was detected positive:
\[Recall=\frac{FP}{FP+TN}=\frac{TP}{P}\]Specificity measures the percentage of the negative population that was detected negative:
\[Specificity=\frac{TN}{TN+FP}=\frac{TN}{N}\]\(F_1\)-Score is an harmonic mean of precision and recall. For two number \(X_1\) and \(X_2\), the harmonic mean is:
\[H(X_1, X_2)=2 \times \frac{X_1 X_2}{X_1 + X_2}\]So the \(F_1\)-Score is:
\[F_1=2 \times \frac{Precision \times Recall}{Precision + Recall}\]Accuracy which is a natural metric is just the percentage of well predict samples:
\[Accuracy = \frac{TP + TN}{TP + TN + FP + FN} = \frac{TP + TN}{n}\]Where:
ROC (for Receiver operating characteristic) Curve is a curve created where each point corresponds to the results obtained for a given threshold. It plots, for every thresholds, the True Positive Rate against the False Positive Rate:

For a threshold of 0, the TPR would be 1 (every element of the positive population detected positive) and the FPR would also be 1 (every element of the negative population detected positive).
For a threshold of 1, the TPR would be 0 (every element of the positive population detected negative) and the FPR would also be 0 (no element of the negative population detected positive).
Other threshold are in between. A perfect classifier would have a TPR of 1 every element of the positive population detected positive) and a FPR of 0 (no element of the negative population detected positive).
AUC for Area Under Curve is the area under the ROC curve or its integral. When using normalized units, AUC is equal to the probability that a classifier will rank a randomly chosen positive instance higher than a randomly chosen negative one.
AUC is related to the Mann–Whitney U and to the Gini coefficient (not the Gini impurity).
See the paragraph dedicated to AUC on the Wikipedia page for ROC Curve.
For all of the classification metrics see:
Most of unsupervised metrics (without labels) are based on the variance intra clusters and the variance inter cluster.
Silhouette coefficient is a clustering metric defined for a single cluster as:
\[s=\frac{b-a}{\max(a,b)}\]Where:
For a set of cluster in is then:
\[s=\frac{1}{n_{clusters}}\sum_{i=1}^{n_{clusters}}\frac{b_i-a_i}{\max(a_i,b_i)}\]If a cluster is very dense and far from its nearest neighbours then is silhouette coefficient will be high. On the contrary a sparse cluster not isolated from its neighbours will have a low silhouette coefficient.
Calinski-Harabasz Index is a clustering metric defined, for a dataset \(E\) as:
\[s=\frac{B}{W}\frac{n_E-k}{k-1}\]Where \(B_k\) is the between group dispersion measure and \(W_k\) is the within-cluster dispersion measure defined by:
With:
The Calinski-Harabasz index is thus the ratio of the sum of between-clusters dispersion (variance inter) and of within-cluster dispersion (variance intra) for all clusters (where dispersion is defined as the sum of distances squared): The \(\frac{n_E-k}{k-1}\) is a penalty on the number of cluster.
For Calinski-Harabasz Index, an higher score is better.
Davies-Bouldin Index is a clustering metric defined, for a dataset \(E\) as:
\[DB=\frac{1}{k}\sum_{i=1}^k \max_{i \ne j}R_{ij}\]Where:
With:
By taking, for each \(i\), the maximum score \(R_{ij}\) the Davies-Bouldin Index just looks at the score for cluster \(i\) compare to its closest neighbour (similar to Silhouette score). It will compare this distance to the sum of the average distance in cluster \(i\) and in cluster \(j\).
Zero is the lowest possible score. Values closer to zero indicate a better partition.
See:
See: